COVID-19 Drugs and Vaccines¶
Last Updated April 16th, 2020
Abstract¶
This is my attempt to use both text and data mining techniques to build a comprehensive network of relationships between a complex and vast swathe of biological components. I use the CORD-19 corpus as well as MEDLINE.
Approach¶
I use a version of NLP that supports real-time entity assisted search and navigation of the MEDLINE repository while simultaneously using PubMed as the underlying search engine. This version is called TPX. TPX is a modular and versatile text-mininig framework designed for biomedical research. I am particularly using a pipeline called PRIORI-T which is prioritized for phenotype-driven rare diseases. It functions by re-purposing specific modules of the TPX framework. These modules include:
- Dictionary Curation
- Annotator for entity annotations
- MEDLINE processor
- Network Creation module; functions to build a heterogenous network of the correlations extracted by the Correlation Extraction module
- Network Augmentation module; augments the network with novel links inferred from a graphic convolutional approach
This is how I re-purposed it for CORD-19:
- I took the entirety of the CORD19 dataset corpus and filtered it for unique full-text articles, using the Corpus Processor module. I also included the complete MEDLINE abstracts which I used the network augmentation module for in order to augment the network with inferred novel connections.
- I used the Annotator module to annotate the corpus based on the following dictionaries: HUMAN_GENE, GENE_SARS, GENE_MERS, GENE_COVID, PHENOTYPE, CHEMICALS, DRUGS, DISEASE, SYMPTOM, GOPROC, GOFUNC, GOLOC, CELLTYPE, TISSUE, ANATOMY, ORGANISM, COUNTRIES, ETHICS TERMS (general terms related to human ethics), NON-PHARMA INTERVENTION (terms related to non-pharmaceutical intervention), SURVEILLANCE TERMS (general terms related to disease surveillance), VACCINE TERMS, VIROLOGY TERMS (general terms used in virology studies) and EARTH SCIENCE TERMS.
- I used the Correlation Extraction module to extract out correlations among these types which are then used by the Network Creation module to build a network that can be queried to obtain information from the corpus.
Ultimatley, this will serve as a knowledge base that can help the COVID-19 research community to obtain useful insights spanning several complex entity types via a combination of text-mining and network analyses.
PyVis Visualization¶
I used PyVis to construct and visualize an intuitive and interactive exploration of this system. Here's a schematic that outlines the overall approach:
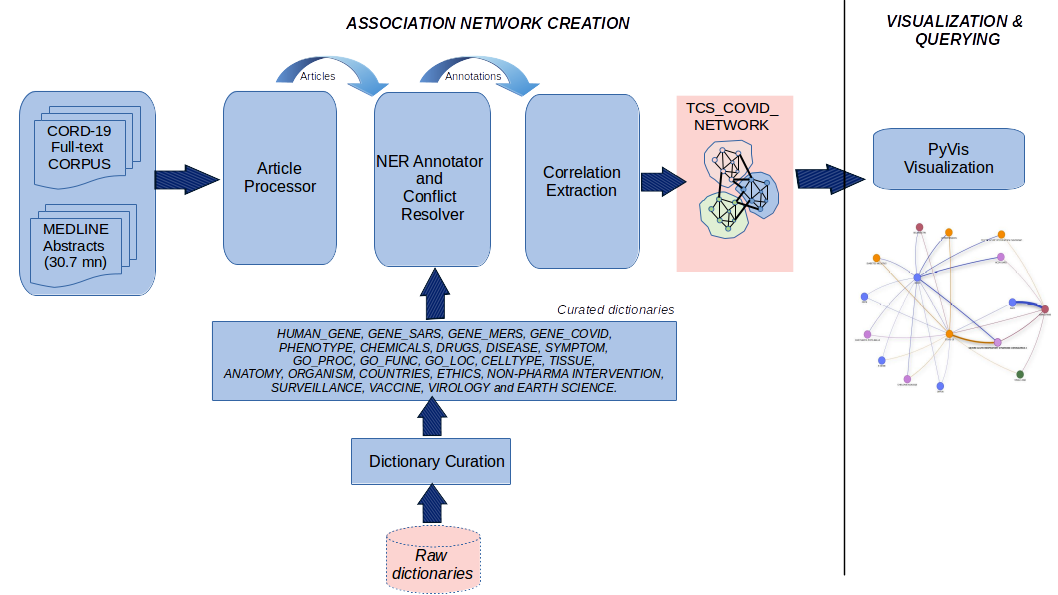
Step-wise Details¶
Corpus preprocessing and augmentation¶
CORD-19 corpus preprocessing As part of the corpus-preprocessing I removed unwanted characters such as white spaces and html codes. I de-duplicated the corpus to remove duplicate articles.
Incorrectly annotated COVID19 articles:: I found this recurring sentence "publicly funded repositories, such as the WHO COVID database with rights for unrestricted research re-use and analyses in any form or by any means with acknowledgement of the original source. These permissions are granted for free by Elsevier for as long as the COVID-19 resource centre remains active." at the begining of 380 articles sourced from Elsevier in the corpus. Many of these articles are actually not relevant to COVID-19 and seem to have come into the corpus because of this one sentence. In such articles, there is only a single mention of COVID-19 in the above sentence. I removed this sentence from the articles.
Instead of focusing only on the "title" and "abstract" sections, I process the COMPLETE corpus of full-text articles. Only Foreign Language Articles were excluded from the corpus. The CORD-19 corpus contains full-text foreign language articles, including those in Spanish, French and German. While these did not adversely affect the overall quality of my system, a few false-positives were observed. Hence, we excluded a total of 854 such foreign language articles.
After the clean-up, I segregated the article text into various spans, which included sentence, section, paragraph and the full article.
Augmenting the corpus with related MEDLINE abstracts: While the provided CORD-19 corpus is an excellent source of COVID-19 articles, I experimented to see if there were any related MEDLINE abstracts that could possibly be added to the corpus to improve the precision and recall. Towards this, I filtered the entire tagged MEDLINE for abstracts containing the terms "coronavirus infection", SARS, MERS or COVID-19 or their synonyms. As a result, I identified a subset of MEDLINE abstracts that could be used to augment the CORD-19 corpus. To reiterate: the provided CORD-19 corpus is the principal data source for my system. However, I did find additional useful COVID-19 related abstracts in MEDLINE.
NER Annotator¶
I used a lexicon (dictionary)-based NER annotator to tag the text. The annotator takes care of local abbreviations.
After tagging, I used the Conflict Resolver to resolve conflicts arising from the same text getting annotated by different dictionaries. The order followed was "DISEASE" > "SYMPTOM" > "PHENOTYPE" > "HGNC" > "GENE-COVID" >"GENE-SARS" >"GENE-MERS" > "VACCINETERMS" > "DRUGS" > "CHEMICALS" > "GO_PROCESS" > "GO_FUNCTION" > "GO_LOCATION" > "CELLTYPE" > "TISSUE" > "ANATOMY" > "VIROLOGYTERMS" > "NONPHARMAINTERVENTIO" > "SURVEILLANCETERMS" > "ETHICSTERMS" > "COUNTRIES" > "EARTHSCIENCETERMS"
Correlation Extraction¶
The Correlation Extraction module uses the Pearson correlation coefficient to compute pairwise correlations between entities identified by the Annotator. To compute the pair-wise correlation for a pair, I used the standard Pearson correlation expression. These probability values are estimated from the corpus. I experimented with computing pair-wise correlations using different text spans: sentence, paragraph, section and article. Based on my evaluation, I found that the correlations computed at the paragraph level were best at striking a balance between precision and recall. I used these associations to build the basic system. Here, the biological entities are the nodes and the pair-wise correlations are represented using undirected weighted edges where the edge weight corresponds to the correlation strength.
Network Augmentation¶
The correlation-based network was further augmented with additional inferred novel links using a graph convolution based technique. These links have no co-occurrence-based support from the corpus. However, such novel links are equally crucial in identifying important drugs, genes etc.,for the drug development, repurposing and vaccine development tasks. I had previously developed a graph convolution-based association inferencing algorithm GCAS (Graph Convolution based Association Scoring) in the context of network-based study of rare-diseases. My approach was motivated by the recent progress in spectral graph convolutions, where association inferencing is viewed as a multi-step signal propagation over the network by convolving the network with a filter.
One of the primary goals of this project is to provide a high quality and a user-friendly network-based abstraction of the given corpus to explore interesting connections between several entity types. However, capturing novel connections are equally crucial for identifying useful targets for drugs and vaccines. We believe that to achieve this, we need to go beyond the provided corpus and consider the complete MEDLINE and other data sources. Our network augmentation task is a step in this direction. My final framework would thus capture highly relevant relations from COVID-19 related corpus and would also mine for additional novel associations from the whole of MEDLINE. Both these knowledge bases are then combined to complement each other so as to amplify the signal and to reduce the overall noise.
Overall, I have combined these knowledge bases (networks) in a simple manner. However, future submissions would provide enhancements to improve the overall quality.
RESULTS¶
I now describe how this network can be used to answer some of the questions posed in the challenge using a network exploration and PyVis visualization approach.
from pyvis.network import Network
import networkx as nx
import pandas as pd
from IPython.display import IFrame
DATA_DIR='./COVID19/CORD19/submissions/'
NETWORK_FILE = DATA_DIR+'cord19_medline_assocs_v2.3.tsv'
ENTITY_NAME_FILE = DATA_DIR+'dicts/entity_name.tsv'
ENT_METADATA_FILE = DATA_DIR+'dicts/entity_metadata.csv'
ent_name=None
ent_id_name=None
ent_name_id={}
enttype_map=None
ent_cmap=None
ent_srcmap=None
connected_nodes=False
notebook_mode=True
net_options = {
"nodes": {
"scaling": {
"min": 46
}
},
"edges": {
"color": {
"inherit": True
},
"shadow": {
"enabled": True
},
"smooth": True
},
"interaction": {
"hover": True,
"navigationButtons": True
},
"physics": {
"enabled": True,
"forceAtlas2Based": {
"gravitationalConstant": -208,
"springLength": 150
},
"minVelocity": 0.05,
"timestep":0.1,
"solver": "forceAtlas2Based"
}
}
def getEntityMaps():
ent_meta_map = pd.read_csv(ENT_METADATA_FILE, sep=',')
enttype_map = ent_meta_map[['entid','enttype','entsource']]
ent_name_df = pd.read_csv(ENTITY_NAME_FILE, sep='\t', converters={'TypeId':str})
ent_name_df.TypeId=ent_name_df.TypeId.str.upper()
ent_name_df.Synonym=ent_name_df.Synonym.str.upper()
ent_name_df.DictId = ent_name_df.DictId.map(enttype_map.set_index('entid')['enttype'])
ent_name = ent_name_df.set_index('Synonym').to_dict()
ent_id_name = ent_name_df.set_index(['TypeId','DictId']).to_dict()
ent_color = ent_meta_map[['enttype','entcolor']]
ent_cmap = ent_color.set_index('enttype').to_dict(orient='index')
ent_source = ent_meta_map[['enttype','entsource']]
ent_srcmap = ent_source.set_index('enttype').to_dict(orient='index')
return enttype_map, ent_name, ent_id_name, ent_cmap, ent_srcmap
def getNetwork():
nw = pd.read_csv(NETWORK_FILE,sep='\t',converters={'src_ent':str, 'target_ent':str})
nw.src_type = nw.src_type.map(enttype_map.set_index('entid')['enttype'])
nw.target_type = nw.target_type.map(enttype_map.set_index('entid')['enttype'])
nw.src_ent=nw.src_ent.str.upper()
nw.target_ent=nw.target_ent.str.upper()
return nw
def buildQueryCriteria(src_ents, source_ent_types=None, target_ents=None, target_ent_types=None,
queryByEntityName=True, topk=50, topkByType=None, connected_nodes=False,
indirect_links=None, inference=False):
# Normalize it upper-case
src_ents = [i.upper() for i in src_ents]
criteria = {'src_ents' : src_ents,
'src_ent_types': source_ent_types,
'target_ents': target_ents,
'target_ent_types': target_ent_types,
'query_entname' : queryByEntityName,
'topk' : topk,
'topkByType' : topkByType,
'connected_nodes' : connected_nodes,
'indirect_links' : indirect_links,
'inference':inference
}
return criteria
def queryByEntityTypes(nw, src_ent_types, target_ent_types):
#Fetch the network
qnw=None
if(src_ent_types is not None):
qnw = nw[nw.src_type.isin(src_ent_types)]
if(target_ent_types is not None):
qnw = nw[nw.target_type.isin(target_ent_types)]
if(qnw is None):
qnw=nw
return qnw
def queryByEntityID(nw, src_ents, target_ents=None):
#Fetch the network
qnw = nw[nw.src_ent.isin(src_ents)]
if(target_ents is not None):
target_ents = [i.upper() for i in target_ents]
qnw = nw[nw.target_ent.isin(target_ents)]
return qnw
def getEntityIds(ents):
# Normalize it upper-case
ents = [i.upper() for i in ents]
print(' Querying by Entity Name ..')
#Get the entity triplet for the entities
typeids = [ent_name['TypeId'][i] for i in ents]
dictids = [ent_name['DictId'][i] for i in ents]
#Update Entity Name to Entity/Node ID for reference
for i in range(len(ents)):
ent_name_id[ents[i]]=getNodeID(typeids[i], dictids[i])
return typeids, dictids
def queryByEntityName(nw, src_ents, target_ents=None):
typeids, dictids = getEntityIds(src_ents)
qnw = nw[nw.src_ent.isin(typeids) & (nw.src_type.isin(dictids))]
if(target_ents is not None):
target_ents = [i.upper() for i in target_ents]
typeids, dictids = getEntityIds(target_ents)
qnw = nw[nw.src_ent.isin(typeids) & (nw.src_type.isin(dictids))]
return qnw
def queryTopk(nw, topk, topkByType):
qnw=None
if(topkByType!=None):
qnw = nw.groupby(['src_ent','target_type']).head(topkByType)
else:
qnw = nw.groupby(['src_ent','target_type']).head(topk)
return qnw
def queryInferredEdges(nw):
qnw = nw[nw.debug=='I']
return qnw
def queryNetwork(nw, criteria):
# Use the criteria to query the network by entity name
qnw=None
if(criteria['query_entname']==True):
qnw = queryByEntityName(nw, criteria['src_ents'], target_ents=criteria['target_ents'])
else:
qnw = queryByEntityID(nw, criteria['src_ents'], target_ents=criteria['target_ents'])
# Display only inferred edges
if(criteria['inference']==True):
qnw = queryInferredEdges(qnw)
# Query by entity types
qnw = queryByEntityTypes(qnw, criteria['src_ent_types'], criteria['target_ent_types'])
# Display only Top-k entites
qnw = queryTopk(qnw, criteria['topk'], criteria['topkByType'])
return qnw
def getEntityNames(src, target):
src_name = ent_id_name['Synonym'][src]
target_name = ent_id_name['Synonym'][target]
return src_name, target_name
def getNodeID(typeid, dictid):
return typeid+'-'+dictid[:2]
def buildNodeAttributes(e):
# Build Node attributes - node_id, node_label, node_title, node_color
src_label, target_label = getEntityNames((e[0],e[1]), (e[2],e[3]))
# Build src node
src_id = getNodeID(e[0], e[1])
src_title="<b>"+src_label+"</b><br><i>"+e[1]+"<br>"+e[0]+"</i><br>"+ent_srcmap[e[1]]['entsource']
src_color=ent_cmap[e[1]]['entcolor']
# Build target node
target_id = getNodeID(e[2], e[3])
target_title="<b>"+target_label+"</b><br><i>"+e[3]+"<br>"+e[2]+"</i><br>"+ent_srcmap[e[3]]['entsource']
target_color=ent_cmap[e[3]]['entcolor']
return (src_id, src_label, src_title, src_color), (target_id, target_label, target_title, target_color)
def edgeAttributes(ent1, ent2, edge_props):
#Build edge attributes
edge_title = '<b>'+ent1+' --- '+ent2+'</b><br>Article Evidence(s) :<br>'
if('I' in edge_props):
num_arts=0
edge_title+='<b>Inferred from GCAS </b></i>'
else:
edge_prop_arr = edge_props.split(sep=',')
num_arts = int(edge_prop_arr[0])-3
art_type=''
for i in range(3, len(edge_prop_arr)):
art=edge_prop_arr[i].replace("[","")
art=art.replace("]","")
if("FT_" in art):
art=art.replace("FT_","")
art_type='CORD_UID :'
else:
art_type='PUBMED_ID :'
edge_title+=art_type+'<i>'+art+'</i><br>'
if(num_arts>5):
edge_title+='and <i><b>'+str(num_arts)+'</b> more articles ...</i>'
return edge_title
def buildGraph(G, filters=False):
#Define Network layout
net = Network(height="750px", width="100%", bgcolor="white", font_color="black", notebook=notebook_mode)
net.options=net_options
#Convert networkx G to pyvis network
edges = G.edges(data=True)
nodes = G.nodes(data=True)
if len(edges) > 0:
for e in edges:
snode_attr=nodes[e[0]]
tnode_attr=nodes[e[1]]
net.add_node(e[0], snode_attr['label'], title=snode_attr['title'], color=snode_attr['color'])
net.add_node(e[1], tnode_attr['label'], title=tnode_attr['title'], color=tnode_attr['color'])
net.add_edge(e[0], e[1], value=e[2]['value'], title=e[2]['title'])
return net
def applyGraphFilters(G, criteria):
fnodes={}
# Filter1 - Connected nodes
if(criteria['connected_nodes']):
bic = nx.biconnected_components(G)
for i in bic:
if(len(i)>2):
fnodes=i.union(fnodes)
# Get the sub-graph after applying the filter(s)
G=G.subgraph(fnodes)
# Filter2 - 'indirect_links'
il_dicts = criteria['indirect_links']
if(il_dicts is not None):
snode = il_dicts['source_node'] if ('source_node' in il_dicts) else criteria['src_ents'][0]
snode = ent_name_id[snode.upper()]
#Depth=Hops+1
depth=(il_dicts['hops']+1) if('hops' in il_dicts) else 2
if('target_nodes' in il_dicts):
tnodes = il_dicts['target_nodes']
elif(criteria['target_ents'] is not None):
tnodes = criteria['target_ents']
else:
tnodes=criteria['src_ents']
tnodes = [ ent_name_id[i.upper()] for i in tnodes]
# Traverse k-hops from source to target nodes.
paths_between_generator = nx.all_simple_paths(G, source=snode, target=tnodes, cutoff=depth)
indirect_paths=[]
i=0
for k, path in enumerate(paths_between_generator):
#if(len(path)==depth+1):
ce=[]
#print(path)
for j, e in enumerate(path):
if j+1 <= len(path)-1:
ce.append((path[j], path[j+1]))
indirect_paths.extend(ce)
G=G.edge_subgraph(indirect_paths)
return G
def run(criteria):
# Load the entire network
nw_df = getNetwork()
# Query the network with the defined search criteria
qnw = queryNetwork(nw_df, criteria)
# Build association network using the query result
sources = qnw['src_ent']
source_types=qnw['src_type']
targets = qnw['target_ent']
target_types=qnw['target_type']
weights = qnw['score']
stats = qnw['debug']
edge_data = zip(sources, source_types, targets, target_types, weights, stats)
G=nx.Graph()
for e in edge_data:
snode, tnode = buildNodeAttributes(e)
G.add_node(snode[0], label=snode[1], title=snode[2], color=snode[3])
G.add_node(tnode[0], label=tnode[1], title=tnode[2], color=tnode[3])
G.add_edge(snode[0], tnode[0], value=e[4], title=edgeAttributes(snode[1],tnode[1], e[5]))
applyFilter = (criteria['connected_nodes'] or criteria['indirect_links'])
if(applyFilter):
G=applyGraphFilters(G, criteria)
net = buildGraph(G, applyFilter)
if(criteria['inference']==True):
net.options['edges']['dashes']=True
else:
net.options['edges']['dashes']=False
print(' Number of Associations in the Network -->'+str(len(G.edges())))
return net
# Prepare Entity Maps
enttype_map, ent_name, ent_id_name, ent_cmap, ent_srcmap = getEntityMaps()
#Display Entity Types
enttype_map
QueryTerms=['C000657245']
criteria = buildQueryCriteria(QueryTerms, topkByType=15, queryByEntityName=False)
criteria
net = run(criteria)
net.show("cord19_ex1.html")
QueryTerms=['C000657245']
criteria = buildQueryCriteria(QueryTerms,target_ent_types=['DRUGS','CHEMICALS'], topkByType=15, queryByEntityName=False)
criteria
net = run(criteria)
net.show("cord19_ex2.html")
From this network, let's look at some of the chemicals and drugs:¶
Tocilizumab: Supported by PMID:32240462, CORD_UID:yy7abob9, PMID:32241792, PMID:32222713, PMID:32209313 and PMID:32243501
- PMID:32209313: New therapeutic opportunities for COVID-19 patients with Tocilizumab: Possible correlation of interleukin-6 receptor inhibitors with osteonecrosis of the jaw
- PMID:32240462: Coronavirus disease 2019 (COVID-19): a clinical update. The efficacy of some promising antivirals, convalescent plasma transfusion, and tocilizumab needs to be investigated by ongoing clinical trials.
Arbidol: Supported by PMID:32147628, PMID:32171872, CORD_UID:zwqci59h, CORD_UID:5pfusktn, PMID:32037389, CORD_UID:7e8zlt3t
- PMID:32147628 Several drugs such as chloroquine, arbidol, remdesivir, and favipiravir are currently undergoing clinical studies to test their efficacy and safety in the treatment of coronavirus disease 2019 (COVID-19) in China; some promising results have been achieved thus far.
Convalescent plasma: Supported by PMID:32240549, PMID:32219429, PMID:32240462, PMID:32219428, PMID:32220178, PMID:32240545
- PMID:32219428 In this preliminary uncontrolled case series of 5 critically ill patients with COVID-19 and ARDS, administration of convalescent plasma containing neutralizing antibody was followed by improvement in their clinical status.
- PMID:32240549 The recent Coronavirus Disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has prompted not only a search for effective anti-viral treatment and spread control measures, but also a reconsideration of the use of convalescent plasma for COVID-19 treatment [5, 6].
Chloroquine: Supported by PMID:32219882, PMID:32217113, PMID:32219357, PMID:32074550, PMID:32179150, PMID:32212513
- PMID:32179150 Chloroquine, remdesivir, lopinavir, ribavirin or ritonavir have shown efficacy to inhibit coronavirus in vitro.
Prominent Alternative Medicine Chemicals and Drugs
- Shufeng jiedu, supported by articles CORD_UID:0xhho1sh, CORD_UID:k2ixwz9w, CORD_UID:msohf5oa, PMID:32037389, CORD_UID:ptnmtvzj and CORD_UID:athjtu2j
- Lianhuaqingwen, supported by CORD_UID:szsb1oan, CORD_UID:bep0xtxa, PMID:31996494, PMID:32205232, CORD_UID:x23ej29m and CORD_UID:h72w22rm
QueryTerms=['naproxen', 'clarithromycin', 'minocycline', 'covid-19']
criteria = buildQueryCriteria(QueryTerms, topkByType=15)
criteria
net = run(criteria)
net.show("cord19_ex3.html")
Multi-hop queries¶
Neighbourhood network constructed based on the query with entity terms (baricitinib, covid-19 and ace2) and with three additional filters, namely,
- Only entity types ( 'HGNC','DRUGS','GENE_SARS-CoV-2','ORGANISM', 'CHEMICALS', 'VIROLOGY TERMS', 'DISEASE'),
- For each query entity, restrict it's neighbourhood to the Top-15 neighbours for each of the filtered entity types and
- Retain only those entities in the neighbourhood that appear in a multi-hop (simple) path (no more than 3 hops) from 'covid-19' to 'baricitinib'.
QueryTerms=['naproxen', 'clarithromycin', 'minocycline', 'covid-19']
criteria = buildQueryCriteria(QueryTerms, topkByType=15, connected_nodes=True)
criteria
net = run(criteria)
net.show("cord19_ex4.html")
Using Sub-Networks to understand Drug mechanisms and Disease action¶
I now show the utility of connections between different entity types using sub-networks. These could provide a deeper understanding of drug mechanisms, disease action, etc. An important caveat when analyzing such sub-networks - each edge or connection was derived from a particular paragraph of the article(s) in the corpus - hence the correlation is only in that context.
Let's take the example where we look for connections between the drug azithromycin, non-structural proteins - Nsp8, Nsp15, human mitochondrial ribosomal protein S27 (MRPS27) and their possible role in viral repliction/viral genome replication:
QueryTerms=['covid-19']
criteria = buildQueryCriteria(QueryTerms,target_ent_types=['DISEASE','DRUGS'], topkByType=15)
criteria
net = run(criteria)
net.show("cord19_ex5.html")
Correlations from the full-text article "38d6gb7o" from the CORD-19 corpus show:
- Azithromycin---COVID-19 "Similarly, antibiotics like azithromycin have also been mooted as treatments for COVID-19." "While this too remains to be demonstrated, we note that Azithromycin has off-target activity against human mitochondrial ribosomes, components of which interact with the SARS-CoV-2 Nsp8 protein (MRPS5, MRPS27, MRPS2, and MRPS25)."
- This second sentence also gives us Azithromycin---mitochondrial ribosomes, Azithromycin---MRPS27 and Nsp8---MRPS27 edges
- Finally, viral replication---COVID-19 is got from the paragraph ".................stress granules and host translation shutoff 55 . This functionality seems to benefit viral ....are targeted by several SARS-CoV-2 viral proteins. Interestingly, ... ....stress granules and host translation shutoff. This functionality seems to benefit viral replication, as stress granules are inhibitory to replication of MERS-CoV 56 and other viruses"
A separate search of PubMed results in some abstracts not in the corpus that support the correlations mentioned above:
- From PMID:32229706: "Similarly, two proposed therapeutics for the treatment of COVID-19 infection are Azithromycin and Quercetin, both drugs with significant senolytic activity. As Azithromycin and Doxycycline are both commonly used antibiotics that inhibit viral replication and IL-6 production, we may want to consider this general class of antibiotics that functionally inhibits cellular protein synthesis as a side-effect, for the treatment and prevention of COVID-19 disease."
- From PMID:30918070: "Replacement of a conserved Lys residue with Ala abolished the in vitro RNA-binding and TATase activities of nsp8 and caused a nonviable phenotype when the corresponding mutation was introduced into the HCoV-229E genome, confirming that these activities are mediated by nsp8 and critical for viral replication. While confirming the critical role of nsp8 in coronavirus replication, the study amends the list of activities mediated by coronavirus nsp8 proteins in the absence of other proteins."
- From PMID:30135128: "We also hypothesize that the primase-like protein Nsp8 and the Nsp7/Nsp8 complex may interact with Nsp15 and affect enzymatic activity. This contributes to the understanding of the association of Nsp15 with the viral replication and transcription machinery."
QueryTerms=['baricitinib','covid-19', 'ace2']
criteria = buildQueryCriteria(QueryTerms, topkByType=15, target_ent_types=['HGNC','DRUGS','GENE_SARS-CoV-2','ORGANISM', 'CHEMICALS', 'VIROLOGY TERMS', 'DISEASE'],
indirect_links={'source_node':'covid-19', 'target_nodes':['baricitinib'], 'hops':3})
criteria
net = run(criteria)
net.show("cord19_ex6.html")
I now highlight some drugs connected by indirect edges to COVID-19 disease in the network. The caveat here is that since these are not direct connections, there is no reference article that talks about them. One could think of them as putative suggestions! Some text from MEDLINE abstracts about these drugs is also shown.
Ingavirin (pentanedioic acid - imidazolyl ethanamide drug combination)
- PMID:27876718 states "Research objective was to study the efficacy of Ingavirin for prevention of recurrent herpetic stomatitis in employees of Kazan city industrial enterprises frequently suffering from acute respiratory viral infections. The obtained data allow to recommend ingavirin for prevention of recurrent herpetic stomatitis."
- PMID:21033471 states "Despite obvious success in the vaccine development and chemotherapy of influenza, it remains a poorly controlled infection leading to emergence of new pandemic variants of the virus with high morbidity and mortality. We investigated the protective activity of Ingavirin against the lethal influenza A (H1N1) 2009 virus infection on albino mice. Oral use of Ingavirin resulted in sharp decreasing of the mortality (index of protection up to 57%), slight decreasing of the infectious titer of the virus in the lungs ( up to 40-fold), normalizing of the body weight dynamics and the lung tissue structure vs. the placebo-treated control. The degree of the bronchial epithelium damage was also strongly decreased. The results allow to consider Ingavirin as an effective antiviral against the current pandemic influenza virus."
Pocapavir
- PMID:25229269 states "Pocapavir (V-073) is an investigational drug candidate being developed for poliovirus indications, but also has variable antiviral activity against nonpolio enteroviruses. We describe the first use of pocapavir in treating a case of severe neonatal enteroviral sepsis due to Coxsackievirus B3."
R 125489 (laninamivir)
- PMID:28869418 says "Laninamivir octanoate is a recently developed inhaled neuraminidase inhibitor for treating influenza virus infection"
- PMID:30935767 says "Four neuraminidase (NA) inhibitors and an RNA synthesis inhibitor were recently approved and are currently in clinical use for influenza. Among NA inhibitors, oseltamivir phosphate (OSE, Tamiflu®) and zanamivir are approved worldwide, whereas peramivir and laninamivir octanoate (LAN, Inavir®) are regionally approved for human use"
- PMID:22028647 says "The 2009 H1N1 influenza pandemic (pH1N1) led to record sales of neuraminidase ( NA) inhibitors, which has contributed significantly to the recent increase in oseltamivir-resistant viruses. Therefore, development and careful evaluation of novel NA inhibitors is of great interest. Recently, a highly potent NA inhibitor, laninamivir, has been approved for use in Japan."
- A recent article "PMID:32251791, A first Case of Meningitis/Encephalitis associated with SARS-Coronavirus-2"(not in the corpus) mentions Laninamivir in the context of meningitis in a COVID-19 patient.
Peficitinib (an oral Janus Kinase Inhibitor)
- PMID:31093950 says "Peficitinib [Smyraf® (Astellas Pharma)] is a Janus kinase (JAK)1, JAK2, JAK3 and tyrosine kinase (Tyk)2 (pan-JAK) inhibitor recently approved in Japan for the treatment of rheumatoid arthritis. Inhibition of JAK suppresses the activation of cytokine signalling pathways involved in inflammation and joint destruction in rheumatoid arthritis."
- PMID:31181818 mentions "Conclusion: Peficitinib suppressed the JAK- STAT pathway in RA FLS and also suppressed monocyte chemotaxis and proliferation of FLS through inhibition of inflammatory cytokines"
Paritaprevir
- PMID:32108916 says "Elimination of the virus with direct-acting antivirals (DAAs) may modify host immune response via altering these immune checkpoint receptors' expression. We conducted a prospective study to analyze changes in TIM-3, PD-1 and their ligands galectin-9, PD-L1 expression by peripheral blood T cell subpopulations, NK cell subpopulations, and monocytes by multicolor flow cytometry in 14 CHC patients successfully treated with 12 weeks of dasabuvir, ombitasvir, and paritaprevir/ ritonavir plus ribavirin.Our data suggest that DAA treatment not only inhibits viral replication but may alter host adaptive and innate immune responses."
- PMID:28680834 says "DAAs inhibit specific HCV non-structural proteins (NS) that are vital for its replication. Boceprevir, telaprevir, simeprevir, asunaprevir, grazoprevir and paritaprevir are NS3/4A inhibitors."
- In fact, a recent article "PMID:32266873, Targeting SARS-CoV-2: A Systematic Drug Repurposing Approach to Identify Promising Inhibitors Against 3C-like Proteinase and 2'-O-RiboseMethyltransferase."(not in the corpus) mentions Paritaprevir as a potential drug COVID-19, thus showing the potential value of inference!
QueryTerms=['C000657245','D017963', 'NS8_WCPV', 'GO:0019079', 'HGNC:14512']
criteria = buildQueryCriteria(QueryTerms, topkByType=20, queryByEntityName=False, connected_nodes=True)
criteria
net = run(criteria)
net.show("cord19_ex7.html")
QueryTerms=['C000657245']
criteria = buildQueryCriteria(QueryTerms, topkByType=15, queryByEntityName=False, inference=True)
criteria
net = run(criteria)
net.show("cord19_inference.html")
DISCUSSION¶
My contributions are multi-fold:
- Build a pair-wise association network covering a comprehensive set of biomedical entity types extracted from the provided corpus. I also experimented with augmenting the provided corpus with related MEDLINE abstracts.
- Mine for novel inferred associations from the whole MEDLINE and augment the network with these to help in better identification of good drug and vaccine targets.
- Provide an intuitive and interactive graphical exploration of the network using PyVis. I provide a Jupyter notebook for this. To illustrate the utility of the notebook, we have included a set of use cases for exploring the network using PyVis and NetworkX library.
- To answer specific as well as general questions pertinent to this task.
The network captures associations between different entities in the augmented corpus described earlier. The network nodes correspond to the biological entities and the edges correspond to the associations. The edges are weighted where the edge weight denote the strength of association (correlation strength in this network) between the entity pair. The inferred associations from MEDLINE are also included in the network with additional flags. The entities span a comprehensive set of entity types, namely HUMAN_GENE, GENE_SARS, GENE_MERS, GENE_COVID, PHENOTYPE, CHEMICALS, DRUGS, DISEASE, SYMPTOM, GOPROC, GOFUNC, GOLOC, CELLTYPE, TISSUE, ANATOMY, ORGANISM, COUNTRIES, ETHICS TERMS, NON-PHARMA INTERVENTION, SURVEILLANCE TERMS, VACCINE TERMS, VIROLOGY TERMS and EARTH SCIENCE TERMS.
The network is compatible with the NetworkX package. User can identify a network node (biological entity) either by its "term name" or the entity ID in its source DB The ID here is its public ID as present in the public data source from which its source dictionary was built. For instance, the Human Gene dictionary was built using HGNC. Hence, in order to query for "ACE2", one could either search using the the common term name "ACE2" or its HGNC ID "HGNC:13557". We have listed the public data sources for each of the entity types which would help the user to figure out the entity ID/term name to be used for querying. There are few terms in the dictionaries that are inserted manually. These terms can be searched only by the term name. Furthermore, dictionaries for COUNTRIES, ETHICS TERMS, NON-PHARMA INTERVENTION, SURVEILLANCE TERMS, VACCINE TERMS, VIROLOGY TERMS and EARTH SCIENCE TERMS have been built from public sources as well as with some manual updates specifically for this CORD19 challenge. Hence, searching the network for these entity types can be done preferably through querying using term name rather than IDs as few of the network entities have only internal IDs and no public IDs.
An important aspect to be noted is the absence of normalization when building this network, resulting in a potential bias. This is because the provided corpus is highly specific to "COVID-19" disease articles. Though the associations are extracted based on co-occurrences of the entities in these articles, their association strength could be biased due to the limited nature of the corpus. In the subsequent releases, we would be broadening the scope of the network and also be incorporating weight normalizations based on a more comprehensive collection of articles.
In future, associations from high quality and heterogeneous curated data with sufficient coverage can be considered for augmenting the network derived solely from text-mining. These could be from sources such as IntAct. Of course, experiments would need to be done to analyse the effects of addition of such curated associations to text-mined associations in these kinds of networks.